Scientific visualization has moved far beyond static 2D figures and simple line drawings. Today, structural biologists and biochemists can explore and communicate the molecular world using cinematic 3D renderings that combine scientific precision with artistic expression. Among the most powerful tools enabling this transformation are Blender, the open-source 3D graphics suite, together with MolecularNodes, a plugin developed by Brady Johnston to bring molecular data into Blender’s rich, node-based modeling environment.
In this post, we’ll explore how MolecularNodes bridges the gap between raw structural data, such as cif files from the Protein Data Bank (PDB), and professional-grade 3D visualization. We’ll also show how, thanks to Jobs in DECTRIS CLOUD, you can generate a high-quality still image or even a movie render of your molecule with just a few clicks.
Why Use Blender for Scientific Visualization?
Blender is widely recognized in the 3D graphics community for its flexibility, professional rendering engines, and strong Python scripting support. Originally developed for animation and design, Blender’s open architecture has made it a favorite among scientists and educators who want to create compelling scientific visualizations.
Unlike specialized tools such as PyMOL or ChimeraX, which were optimized for molecular analysis, Blender provides total creative freedom. Its powerful rendering engines (Eevee for real-time previews and Cycles for physically accurate ray tracing) make it possible to illuminate molecules with realistic light, depth, and texture.
The main limitation of Blender, until recently, was that it didn’t natively understand molecular data formats like PDB or mmCIF (cif files). Importing atomic coordinates into Blender while preserving biochemical information, including chain identifiers, residue types, or secondary protein structure elements, was cumbersome.
That’s precisely the problem that MolecularNodes solves.
Introducing Brady Johnston’s MolecularNodes
MolecularNodes is an open-source add-on for Blender that allows users to import, manipulate, and render molecular structures directly. Developed by Brady Johnston to serve both scientists and artists, it translates atomic data into Blender’s Geometry Nodes system, which is a procedural modeling framework that represents objects dynamically.
In basic terms: MolecularNodes converts the coordinate data from a PDB or mmCIF file into a manipulable 3D structure, complete with atoms, bonds, ribbons, and molecular surfaces. Because it uses Geometry Nodes, every aspect can be modified interactively and reproducibly.
This combination makes MolecularNodes an ideal bridge between data fidelity and visual storytelling and it is quickly becoming essential in both academic research and science communication. This is especially evident on recent journal covers of Science, Cell and more. Some examples were designed by Verena Resch (Luminous Lab) and can be found here.
Getting Started: From Structure File to Scene
1. Start a Blender Session
You can either download the latest version of Blender (3.x or higher) from https://www.blender.org and add the MolecularNodes plugin.
Or you can start a DECTRIS CLOUD Session with the Blender + MolecularNodes Environment. Everything will be set up for you and as an example, a structure of the ATP Synthase (PDB 6N2Y) will be preloaded.
2. Import a Structure
In the right panel under the Scene menu, you can find the Molecular Nodes section.
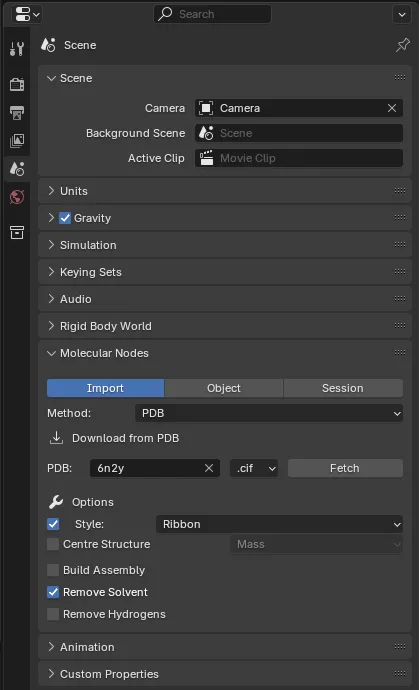
You can load molecular data in two ways:
- From the Protein Data Bank:
- Select PDB in the Method drop-down
- Enter a PDB ID (e.g., 6N2Y)
- Choose a Style (see below) and adjust settings
- Click Fetch.
- From a local file:
- Select Local in the Method drop-down
- Select a .pdb or .cif file on your computer
- Choose a Style (see below) and adjust settings
- Click Local.
MolecularNodes automatically processes the structure, displaying atoms, bonds, or ribbons depending on your chosen representation.
Exploring Molecular Representations
Once loaded, your molecule appears as a Blender object powered by a Geometry Nodes network. If you want to explore different representations designed for scientific or artistic purposes you will have to reload your molecule in one of the following styles:
- Spheres: spheres representing each atom of the molecule.
- Cartoon: smooth surfaces representing the secondary protein structure (α-helices, β-sheets, loops).
- Ribbon: smooth tube through the alpha carbons of the protein.
- Surface: molecular surfaces generated by van der Waals or solvent-excluded models.
- Ball and Stick: cylinders connecting atoms based on bonding rules.
Each representation can be fine-tuned procedurally: color by chain, residue type, or element; adjust sphere sizes or bond thickness; or even blend representations for hybrid views.
This flexibility is invaluable when preparing supplementary material for publications, where visual clarity and data accuracy are equally critical. You can, for instance, highlight ligand binding sites or emphasize conformational changes in a protein’s secondary structure and use Blender’s power to generate impressive camera movements.
Material Design and Shading
In Blender’s Shading workspace, you can modify the look and feel of your molecular model. Each atom, bond, or surface inherits metadata that can drive procedural materials.
This deep integration with Blender’s powerful material system allows you to blend scientific accuracy with artistic style, opening up creative opportunities for figures, teaching materials, or journal cover art.
Lighting and Camera
Lighting plays a crucial role in making your visualization engaging and readable. Blender’s rendering engines, Eevee (real-time) and Cycles (ray-traced), offer full control over illumination and realism.
Setting up a camera to highlight your molecule’s most important features and enabling Depth of Field you can create a soft, microscopy-like focus, drawing the viewer’s eye to specific residues or domains. Blender’s animation system allows you to move your camera allowing you to create movies of your molecular models by rotating them, flying through them, or even showing conformational changes.
One-Click Rendering with DECTRIS CLOUD Jobs
While Blender is powerful, the learning curve is steep. Often, it is sufficient to generate a single high quality render or a turntable-style animation of your molecule.
With DECTRIS CLOUD, you can use the Render Molecule Job Template to create still images and movies of your favourite protein with a single click. You can come back months or even years later to re-run the same render with a different style or a different resolution without having to set anything up again. The reproducibility is built into DECTRIS CLOUD Jobs.
The results are returned to you as downloadable assets or you can directly share them with your friends and collaborators via DECTRIS CLOUD.
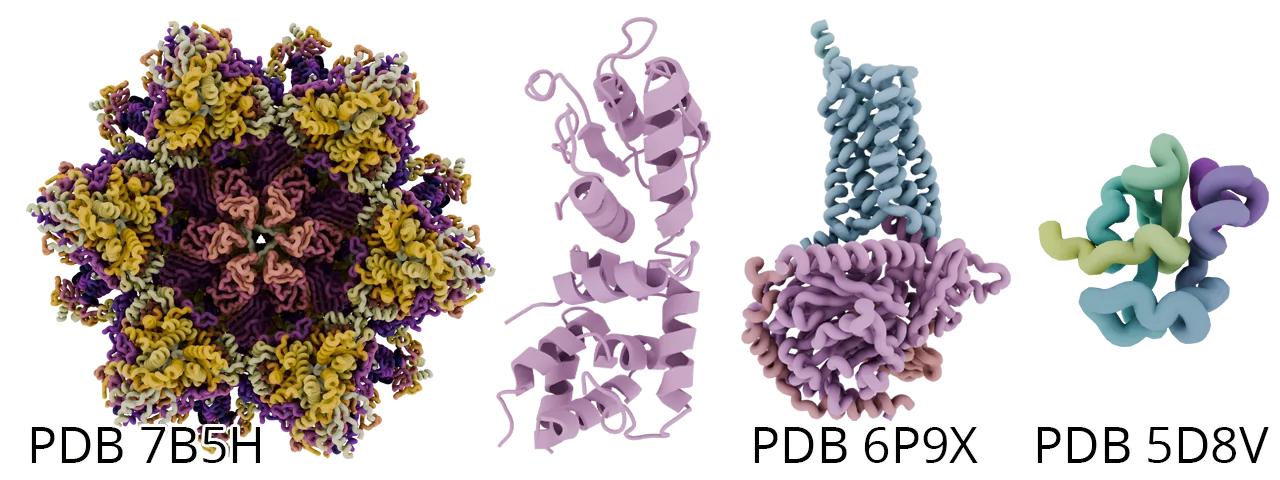
Conclusion
Rendering molecules with Blender and MolecularNodes transforms structural biology data into stunning, interactive art. Whether you’re exploring secondary protein structure, preparing supplementary materials, or creating visuals for a talk or journal cover, these tools let you communicate science with clarity and creativity.
.webp)




